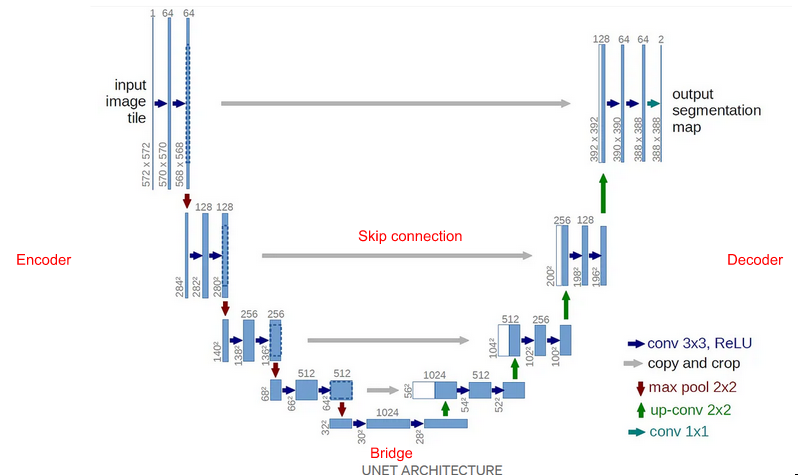
Encoder-Decoder architecture
Components
Encoder
Encoder part halves the spatial dimensions and doubles the number of filters on each block
Each encoder block consists of two 3×3 convolutions, where each convolution is followed by a ReLU (Rectified Linear Unit) activation function
Follows a 2×2 max-pooling, where the spatial dimensions (height and width) of the feature maps are reduced by half
Skip connections
Additional information that helps the decoder to generate better semantic features. T
Helps the indirect flow of gradients to the earlier layers without any degradation.
Bridge
Connects encoder & decoder, consists of two 3×3 convolutions, where each convolution is followed by a ReLU activation function
Decoder
Decoder part doubles the spatial dimension and halves the number of filters in each block.
starts with a 2×2 transpose convolution, concatenated with the corresponding skip connection feature map from the encoder block
Then two 3×3 convolutions are used, where each convolution is followed by a ReLU activation function
Output
The output of the last decoder passes through a 1×1 convolution with sigmoid activation. The sigmoid activation function gives the segmentation mask representing the pixel-wise classification
Architecture Used For Semantic Segmentation

Input shape: (400, 400, 1) (height, width, channels)
Number of filters (n_filters): 16
Dropout rate (dropout): 0.05
Encoding path
Block 1
c1 = conv2d_block(input_img, n_filters * 1, kernel_size = 3, batchnorm = batchnorm)
p1 = MaxPooling2D((2, 2))(c1)
p1 = Dropout(dropout)(p1)
C1
Apply a convolution operation 400 x 400 input image using 16 filters of 3×3
Input: (400, 400, 1)
Output (without padding): (398, 398, 16)
Output (with padding): (400, 400, 16)
P1
Reduce the spatial dimensions of c1 by taking the maximum value in each 2×2 region
Input: (400, 400, 16)
Output: (200, 200, 16)
Dropout
Randomly setting 5% of the values in p1 to zero to prevent overfitting
Input: (200, 200, 16)
Output: (200, 200, 16)
Block 2
c2 = conv2d_block(p1, n_filters * 2, kernel_size = 3, batchnorm = batchnorm)
p2 = MaxPooling2D((2, 2))(c2)
p2 = Dropout(dropout)(p2)
C2
Apply a convolution operation on p1 with 32 filters
Input: (200, 200, 16)
Output: (200, 200, 32)
P2
Reduce the spatial dimensions of c2 using max pooling
Input: (200, 200, 32)
Output: (100, 100, 32)
Dropout
Randomly setting 5% of the values in p2 to zero
Input: (100, 100, 32)
Output: (100, 100, 32)
Block 3
c3 = conv2d_block(p2, n_filters * 4, kernel_size = 3, batchnorm = batchnorm)
p3 = MaxPooling2D((2, 2))(c3)
p3 = Dropout(dropout)(p3)
C3
applying a convolution operation on p2 with 64 filters
Input: (100, 100, 32)
Output: (100, 100, 64)
P3
reducing the spatial dimensions of c3 using max pooling
Input: (100, 100, 64)
Output: (50, 50, 64)
Dropout
Randomly setting 5% of the values in p2 to zero
Input: (50, 50, 64)
Output: (50, 50, 64)
Block 4
c4 = conv2d_block(p3, n_filters * 8, kernel_size = 3, batchnorm = batchnorm)
p4 = MaxPooling2D((2, 2))(c4)
p4 = Dropout(dropout)(p4)
C4
applying a convolution operation on p1 with 128 filters
Input: (50, 50, 64)
Output: (50, 50, 64)
P4
reducing the spatial dimensions of c2 using max pooling
Input: (50, 50, 64)
Output: (25, 25, 64)
Dropout
Randomly setting 5% of the values in p2 to zero
Input: (25, 25, 64)
Output: (25, 25, 64)
Bridge
c5 = conv2d_block(p4, n_filters = n_filters * 16, kernel_size = 3, batchnorm = batchnorm)
High level presentation of image features
Input: (25, 25, 64)
Output: (25, 25, 256)
Decoding path
Block 6
u6 = Conv2DTranspose(n_filters * 8, (3, 3), strides = (2, 2), padding = ‘same’)(c5)
u6 = concatenate([u6, c4])
u6 = Dropout(dropout)(u6)
c6 = conv2d_block(u6, n_filters * 8, kernel_size = 3, batchnorm = batchnorm)
U6 Convolution
applying a transposed convolution operation on c5 with 8 filters ((n_filters*8)/2 = 128), a 3×3 kernel, and a stride of 2×2
Input: (25, 25, 256)
Output: (50, 50, 128)
U6 Concatenation
Concatenate the upsampled features u6 with the corresponding features from the contracting
path c4
Input: (50, 50, 128), (50, 50, 128)
Output: (50, 50, 256)
U6 Dropout
Randomly set 5% of the values in u6 to zero
Input: (50, 50, 256)
Output: (50, 50, 256)
C6
Apply a convolutional block on u6 with (n_filters*8)/2 = 128 filters
Input: (50, 50, 256)
Output: (50, 50, 128)
Block 7
u7 = Conv2DTranspose(n_filters * 4, (3, 3), strides = (2, 2), padding = ‘same’)(c6)
u7 = concatenate([u7, c3])
u7 = Dropout(dropout)(u7)
c7 = conv2d_block(u7, n_filters * 4, kernel_size = 3, batchnorm = batchnorm)
U7 Convolution
applying a transposed convolution operation on c6 with 4 filters i.e. ((n_filters*4)/2 = 64), a 3×3 kernel, and a stride of 2×2
Input: (50, 50, 128)
Output: (100, 100, 64)
U7 Concatenation
Concatenate the upsampled features u7 with the corresponding features from the contracting
path c3
Input: (100, 100, 64)(100, 100, 64)
Output: (100, 100, 128)
U7 Dropout
Randomly set 5% of the values in u6 to zero
Input: (100, 100, 128)
Output: (100, 100, 128)
C7
Apply a convolutional block on u7 with 4 i.e. (32*4=128/2=64)
Input: (100, 100, 128)
Output: (100, 100, 64)
Block 8
u8 = Conv2DTranspose(n_filters * 2, (3, 3), strides = (2, 2), padding = ‘same’)(c7)
u8 = concatenate([u8, c2])
u8 = Dropout(dropout)(u8)
c8 = conv2d_block(u8, n_filters * 2, kernel_size = 3, batchnorm = batchnorm)
U8 Convolution
applying a transposed convolution operation on c7 with 2 filters i.e. ((n_filters*2)/2 = 32), a 3×3 kernel, and a stride of 2×2
Input: (100, 100, 64)
Output: (200, 200, 32)
U8 Concatenation
Concatenate the upsampled features u8 with the corresponding features from the contracting
path c2
Input: (200, 200, 32)(200, 200, 32)
Output: (200, 200, 64)
U8 Dropout
Randomly set 5% of the values in u8 to zero
Input: (200, 200, 64)
Output: (200, 200, 64)
C8
Apply a convolutional block on u8 with 2 filters i.e. ((n_filters*2)/2 = 32)
Input: (200, 200, 64)
Output: (200, 200, 32)
Block 9
u9 = Conv2DTranspose(n_filters * 1, (3, 3), strides = (2, 2), padding = ‘same’)(c8)
u9 = concatenate([u9, c1])
u9 = Dropout(dropout)(u9)
c9 = conv2d_block(u9, n_filters * 1, kernel_size = 3, batchnorm = batchnorm)
U9 Convolution
applying a transposed convolution operation on c8 with filters i.e. ((n_filters*1)/2 = 16), a 3×3 kernel, and a stride of 2×2
Input: (200, 200, 64)
Output: (400, 400, 16)
U9 Concatenation
Concatenate the upsampled features u8 with the corresponding features from the contracting
path c1
Input: (400, 400, 16)(400, 400, 16)
Output: (400, 400, 32)
U9 Dropout
Randomly set 5% of the values in u8 to zero
Input: (400, 400, 32)
Output: (400, 400, 32)
C9 Convolution
Apply a convolutional block on u8 with filters i.e. ((n_filters*1)/2 = 16) filters
Input: (400, 400, 32)
Output: (400, 400, 16)
Output
outputs = Conv2D(1, (1, 1), activation=’sigmoid’)(c9)
Convolution Layer with sigmoid activation
Apply a 1×1 convolution to obtain the final output with a sigmoid activation function
Input: (400, 400, 16)
Output: (400, 400, 1)
model = Model(inputs=[input_img], outputs=[outputs])
Math
Taking an instance of Block 1
C1 2D Convolution
Apply a convolution operation 400 x 400 input image using 16 filters of 3×3
Input: (400, 400, 1)
Output (without padding): (398, 398, 16)
Output (with padding): (400, 400, 16)
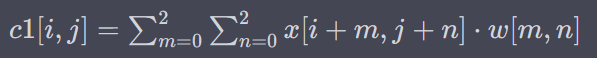
x[i+m, j+n] represents the elements of the input matrix x within the region covered by the convolutional filter, and w[m,n] represents the corresponding weights of the filter
BatchNormalization
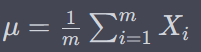
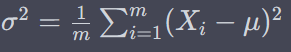
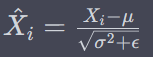
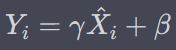
where gamma is scale parameter and beta is shift parameter – learnable*
P1 Max Pooling
Reduce the spatial dimensions of c1 by taking the maximum value in each 2×2 region
Input (with padding): (400, 400, 16)
Output (without padding): (199, 199, 16)
Output (with padding): (200, 200, 16)
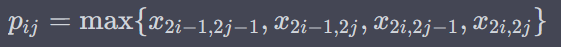
P1 Dropout
Randomly setting 5% of the values in p1 to zero to prevent overfitting

Output of convolution
U6 Transposed 2D convolution
applying a transposed convolution operation on c5 with 8 filters, a 3×3 kernel, and a stride of 2×2
Input: (25, 25, 256)
Output: (50, 50, 128)
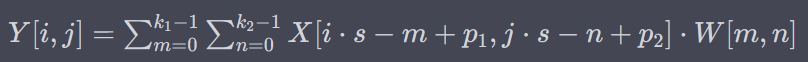
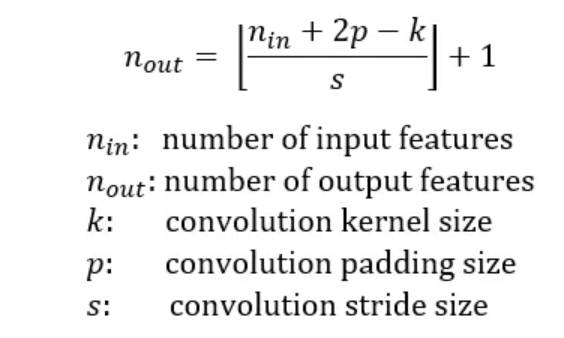
S = stride, p1, p2 = padding
U6 Concatenation
Concatenate the upsampled features u6 with the corresponding features from the contracting
path c4
Input: (50, 50, 128), (50, 50, 128)
Output: (50, 50, 256)
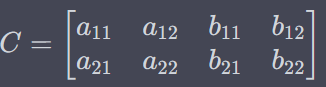
Output
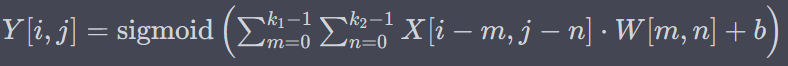
Sigmoid
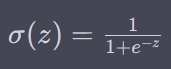
ReLU
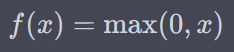
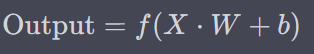
Where f could be either sigmoid or reLu
OTHER
The following are some explanations on the internet that were interesting
Output of convolution
Adam optimizer
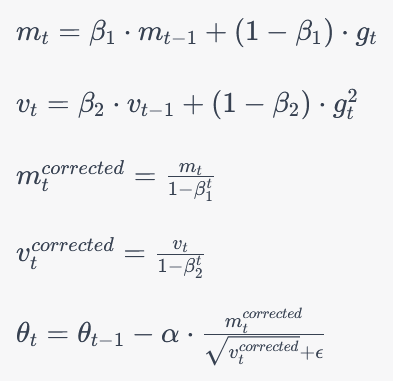
Theta_t = parameter to optimize, alpha = learning rate, beta = exponential decay, m=first order mean estimate of gradient, v = second order uncentered variance of gradient, g = gradient loss wrt parameter, t=time step, epsilon = constant added for stability
References
Components – https://medium.com/analytics-vidhya/what-is-unet-157314c87634
Math – https://towardsdatascience.com/understanding-semantic-segmentation-with-unet-6be4f42d4b47
Other interesting things:
Useful Links:
UNet Line by Line Explanation
